Mammalian Phylogenetics
Cetacean Phylogenetics
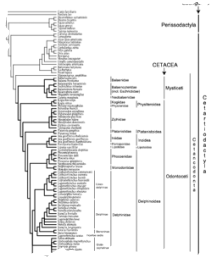
In the mid 1990s cytochrome b and other mitochondrial DNA data reinvigorated cetacean phylogenetics by proposing many novel and provocative hypotheses of cetacean relationships. These results sparked a revision and reanalysis of morphological datasets, and the collection of new nuclear DNA data from numerous loci. Some of the most controversial mitochondrial hypotheses have now become benchmark clades, corroborated with nuclear DNA and morphological data; others have been resolved in favor of more traditional views. That major conflicts in cetacean phylogeny are disappearing is encouraging. However, most recent papers aim specifically to resolve higher-level conflicts by adding characters, at the cost of densely sampling taxa to resolve lower-level relationships. More detailed molecular phylogenies will provide better tools for evolutionary studies. Until more genes are available for a high number of taxa, can we rely on readily available single gene mitochondrial data? In this study, we estimated the phylogeny of 66 cetacean taxa and 24 outgroups based on Cytb sequences. We judged the reliability of our phylogeny based on the recovery of several deep-level benchmark clades. For more details on this study see May-Collado and Agnarsson (2006).
Cetartiodactyla Phylogenetics
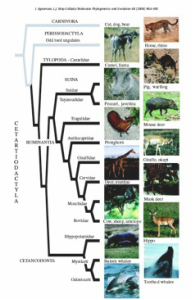
In this study we performed Bayesian phylogenetic analyses on cytochrome b sequences from 264 of the 290 extant cetartiodactyl mammals (whales plus even-toed ungulates) and two recently extinct species, the ‘Mouse Goat’ and the ‘Irish Elk’. Previous primary analyses have included only a small portion of the species diversity within Cetartiodactyla, while a complete supertree analysis lacks resolution and branch lengths limiting its utility for comparative studies. The benefits of using a single-gene approach include rapid phylogenetic estimates for a large number of species. However, single-gene phylogenies often differ dramatically from studies involving multiple datasets suggesting that they often are unreliable. However, based on recovery of benchmark clades—clades supported in prior studies based on multiple independent datasets—and recovery of undisputed traditional taxonomic groups, Cytb performs extraordinarily well in resolving cetartiodactyl phylogeny when taxon sampling is dense. Missing data, however, (taxa with partial sequences) can compromise phylogenetic accuracy, suggesting a tradeoff between the benefits of adding taxa and introducing question marks. In the full data, a few species with a short sequences appear misplaced, however, sequence length alone seems a poor predictor of this phenomenon as other taxa with equally short sequences were not conspicuously misplaced. Although we recommend awaiting a better supported phylogeny based on more character data to reconsider classification and taxonomy within Cetartiodactyla, the new phylogenetic hypotheses provided here represent the currently best available tool for comparative species-level studies within this group. For more details on this work see Agnarsson & May-Collado 2008.
Carnivora Phylogenetics

Phylogenies underpin comparative biology as high-utility tools to test evolutionary and biogeographic hypotheses, inform on conservation strategies, and reveal the age and evolutionary histories of traits and lineages. As tools, most powerful are those phylogenies that contain all, or nearly all, of the taxa of a given group. Despite their obvious utility, such phylogenies, other than summary ‘supertrees’, are currently lacking for most mammalian orders, including the order Carnivora. Carnivora consists of about 270 extant species including most of the world’s large terrestrial predators (e.g., the big cats, wolves, bears), as well as many of man’s favorite wild (panda, cheetah, tiger) and domesticated animals (dog, cat). Distributed globally, carnivores are highly diverse ecologically, having occupied all major habitat types on the planet and being diverse in traits such as sociality, communication, body/brain size, and foraging ecology. Thus, numerous studies continue to address comparative questions within the order, highlighting the need for a detailed species-level phylogeny. Here we present a phylogeny of Carnivora that increases taxon sampling density from 28% in the most detailed primary-data study to date, to 82% containing 243 taxa (222 extant species, 17 subspecies). For more details see Agnarsson et al. 2010.
Afrotherian Phylogenetics

Afrotheria is a recently recognized clade grouping anatomically and biologically diverse placental mammals: elephants and mammoths, dugong and manatees, hyraxes, tenrecs, golden moles, elephant shrews and aardvark. To date, phylogenetic studies have focused on understanding higher level relationships among the major groups within Afrotheria. Here, we provide a species-level phylogeny of Afrotheria based on nine molecular loci, placing nearly 70% of the extant afrotherian species (50) and five extinct species. Our results support the monophyly of Afrotheria and its sister relationship to Xenarthra. Within Afrotheria, the basal division into Afroinsectiphilia (aardvark, tenrecs, golden moles and elephant shrews) and Paenungulata (hyraxes, dugongs, manatees and elephants) is supported, as is the monophyly of all afrotherian families: Elephantidae, Procaviidae, Macroscelididae, Chrysochloridae, Tenrecidae, Trichechidae and Dugongidae. Within Afroinsectiphilia, we recover the most commonly proposed topology (Tubulidentata sister to Afroscoricida plus Macroscelidea). Within Paenungulata, Sirenia is sister to Hyracoidea plus Proboscidea, a controversial relationship supported by morphology. Within Proboscidea, the mastodon is sister to the remaining elephants and the woolly mammoth sister to the Asian elephant, while both living elephant genera, Loxodonta and Elephas are paraphyletic. Top ranking evolutionarily unique species always included the aardvark, followed by several species of elephant shrews and tenrecs. For conservation priorities top ranking species always included the semi-aquatic Nimba otter shrew, some poorly known species, such as the Northern shrew tenrec, web-footed tenrec, giant otter shrew and Giant golden mole, as well as high profile conservation icons like Asian elephant, dugong and the three species of manatee. For more details on this work see Kuntner et al. 2011.
Marsupial phylogeny
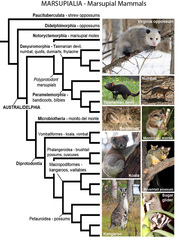
Marsupials or metatherians are a group of mammals that are distinct in giving birth to young at early stages of development and in having a prolonged investment in lactation. The group consists of nearly 350 extant species, including kangaroos, koala, possums, and their relatives. Marsupials are an old lineage thought to have diverged from early therian mammals some 160 million years ago in the Jurassic, and have a remarkable evolutionary and biogeographical history, with extant species restricted to the Americas, mostly South America, and to Australasia. Although the group has been the subject of decades of phylogenetic research, the marsupial tree of life remains controversial, with most studies focusing on only a fraction of the species diversity within the infraclass. Here we present the first Methaterian species-level phylogeny to include 80% of the extant marsupial species and five nuclear and five mitochondrial markers obtained from Genbank and a recently published retroposon matrix. Our primary goal is to provide a summary phylogeny that will serve as a tool for comparative research. We evaluate the extent to which the phylogeny recovers current phylogenetic knowledge based on the recovery of “benchmark clades” from prior studies—unambiguously supported key clades and undisputed traditional taxonomic groups. The Bayesian phylogenetic analyses recovered nearly all benchmark clades but failed to find support for the suborder Phalagiformes. The most significant difference with previous published topologies is the support for Australidelphia as a group containing Microbiotheriidae, nested within American marsupials. However, a likelihood ratio test shows that alternative topologies with monophyletic Australidelphia and Ameridelphia are not significantly different than the preferred tree. Although further data are needed to solidify understanding of Methateria phylogeny, the new phylogenetic hypothesis provided here offers a well resolved and detailed tool for comparative analyses, covering the majority of the known species richness of the group.May-Collado LJ, Kilpatrick CW, Agnarsson I. (2015) Mammals from ‘down under’: a multi-gene species-level phylogeny of marsupial mammals (Mammalia, Metatheria)
PeerJ 3:e805 http://dx.doi.org/10.7717/peerj.805
PeerJ 3:e805 http://dx.doi.org/10.7717/peerj.805
Bat Phylogenetics
Despite their obvious utility, detailed species-level phylogenies are lacking for many groups, including several major mammalian lineages such as bats. Here we provide a cytochrome b genealogy of over 50% of bat species (648 terminal taxa). Based on prior analyzes of related mammal groups, cytb emerges as a particularly reliable phylogenetic marker, and given that our results are broadly congruent with prior knowledge, the phylogeny should be a useful tool for comparative analyzes. Nevertheless, we stress that a single-gene analysis of such a large and old group cannot be interpreted as more than a crude estimate of the bat species tree. Analysis of the full dataset supports the traditional division of bats into macro- and microchiroptera, but not the recently proposed division into Yinpterochiroptera and Yangochiroptera. However, our results only weakly reject the former and strongly support the latter group, and furthermore, a time calibrated analysis of a pruned dataset where most included taxa have the entire 1140bp cytb sequence finds monophyletic Yinpterochiroptera. Most bat families and many higher level groups are supported, however, relationships among families are in general weakly supported, as are many of the deeper nodes of the tree. The exceptions are in most cases apparently due to the misplacement of species with little available data, while in a few cases the results suggest putative problems with current classification, such as the non-monophyly of Mormoopidae. We provide this phylogenetic hypothesis, and an analysis of divergence times, as tools for evolutionary and ecological studies that will be useful until more inclusive studies using multiple loci become available. Agnarsson et al. 2012. http://currents.plos.org/treeoflife/article/a-time-calibrated-species-level-3chrbtx927cxs-5/
